Cold Spring Harbor, New York, USA
February 19, 2025
 Tomato plants mutated with CRISPR, like this one, tended to flower earlier than usual.
Tomato plants mutated with CRISPR, like this one, tended to flower earlier than usual.
Humans have appreciated the beauty of flowers for centuries. Yet, flowers aren’t just aesthetically pleasing. They also play a crucial role in plant reproduction. In all plants, a well-studied gene with a curious name, Unusual Floral Organs (UFO), orchestrates the flowering process. UFO expression hinges on another complex process called cis-regulation. And this one has remained a “black box” of plant biology research for years.
Now, using CRISPR gene editing, Cold Spring Harbor Laboratory (CSHL) Professor and HHMI Investigator Zachary Lippman and his colleagues have begun to uncover the ways in which fragments of non-coding DNA called cis-regulatory sequences dictate how, when, and at what level UFO is expressed. Lippman says this work could one day help researchers make better decisions about which genes to manipulate for more desirable crops. He explains:
“We could have chosen many other genes. We chose this one because it was pretty clear it was going to have that kind of exquisite control. That’s because the flower is a complex structure, and the genes that control its development are very regulated in time, space, and levels.”
The researchers focused on two distantly related flowering plants: tomato and Arabidopsis. First, they identified DNA sequences that don’t code for proteins but are still present in the segment of DNA that turns UFO on and off in both plants.
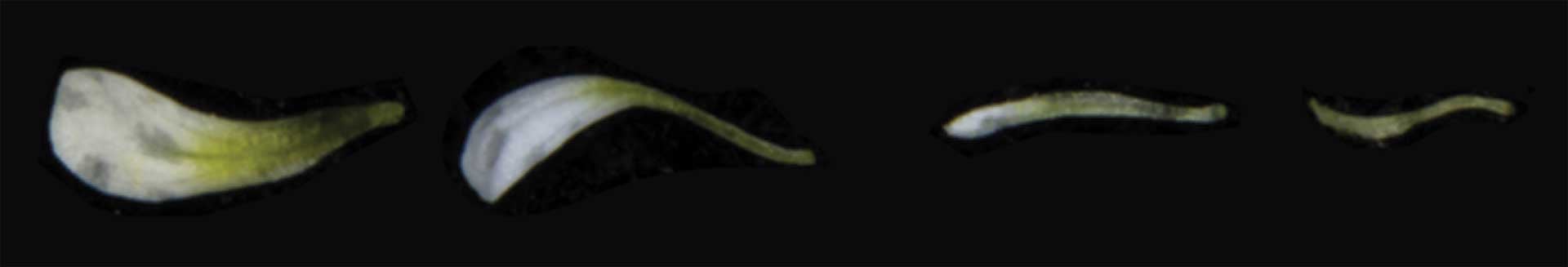 Arabidopsis plants mutated with CRISPR (right) had much smaller flower petals than their wild-grown counterparts (left).
Arabidopsis plants mutated with CRISPR (right) had much smaller flower petals than their wild-grown counterparts (left).
The mere fact the sequences are conserved makes them good candidates for targeting, Lippman says. “It’s a good indication that those sequences have been selected by evolution because they’re important in controlling gene expression.” However, he adds:
“You can’t know until you actually do mutations in those sequences and see what happens.”
Lippman and his team manipulated those non-coding sequences with CRISPR to see how it would affect flower formation. They discovered that the sequences strongly impact flowering in both plants. However, manipulations affect each species differently. For example, deleting a certain sequence in tomatoes resulted in flower formation, but deleting the matching sequence in Arabidopsis suppressed flowering.
“It’s fascinating that different deletions had opposite effects on flowering,” says CSHL postdoc Amy Lanctot. “It seems these sequences act together to balance each other out and make sure plants are flowering at the right place and time.”
The finding could help biologists better understand how cis-regulatory fragments control gene function. “The goal is to reach a better understanding of how functionally complex cis-regulatory DNA is,” Lippman says. “If we can do that, we can better determine which sequences we want to mutate and what kind of mutations we want to make.”
Funding
National Science Foundation, Howard Hughes Medical Institute
Citation
Lanctot, A., et al., “Antagonizing cis-regulatory elements of a conserved flowering gene mediate developmental robustness”, PNAS, February 18, 2025. DOI: 10.1073/pnas.2421990122