Berkeley, California
January 29, 2009
Taking a hint from the text
comparison methods used to detect plagiarism in books, college
papers and computer programs,
University of California, Berkeley researchers have
developed an improved method for comparing whole genome
sequences.
With nearly a thousand genomes partly or fully sequenced,
scientists are jumping on comparative genomics as a way to
construct evolutionary trees, trace disease susceptibility in
populations, and even track down people's ancestry.
To date, the most common techniques have relied on comparing a
limited number of highly conserved genes - no more than a couple
dozen - in organisms that have all these genes in common.
The new method can be used to compare even distantly related
organisms or organisms with genomes of vastly different sizes
and diversity, and can compare the entire genome, not just a
selected small fraction of the gene-containing portion known to
code for proteins, which in the human genome is only 1 percent
of the DNA.
The technique produces groupings of organisms largely consistent
with current groupings, but with some interesting discrepancies,
according to Sung-Hou Kim, professor of chemistry at UC Berkeley
and faculty researcher at Lawrence Berkeley National Laboratory.
However, the relative positions of the groups in the family tree
- that is, how recently these groups evolved - are quite
different from those based on conventional gene alignment
methods.
The computational results have surprised scientists in being
able to classify some bacteria and viruses that until now were
enigmatic.
The technique, which employs feature frequency profiles (FFP),
is described in a paper to appear this week in the early online
edition of the journal Proceedings of the National Academy of
Sciences.
Whole-genome vs. gene-centric methods
Current methods for comparing the genomes of different organisms
focus on a small set of genes that the organisms being compared
have in common. The genomes are then lined up in order to count
the sequence similarities and differences, from which a computer
program constructs a family tree, with near relatives assumed to
have more similar sequences than distant relatives.
This technique assumes organisms have genes in common, however,
or that these "homologous" genes can be identified. When
comparing distantly related species - such as bacteria that live
in vastly different environments - this gene-centric method may
not work, Kim said.
"What do you do when one gene tells you the organisms are
closely related, and another gene tells you they're distantly
related?" he asked. "It happens."
Kim, who in the past focused on creating three-dimensional
demographic maps of all known protein structures, wanted a
technique that could be used to compare genomes of all sizes,
and even genomes only partially sequenced. He also wanted a
method that would compare all regions of the genome, not just
the exons - that is, the DNA transcribed into mRNA, the
blueprint for proteins. Exons make up only 1 percent of the
human genome, with the remainder being non-coding "introns,"
regulatory DNA, duplicate or redundant DNA and transposons -
genes that have jumped from other places in the genome.
Kim thought that traditional text comparison - used, for
example, to assess the authorship of a work of literature or to
identify plagiarized text - might provide a model for whole
genome comparison and a way to test comparison methods. But
while text comparison involves looking at word frequency;
genomes cannot be broken down into words.
"I can compare two books in two different ways. I can pick a few
sentences, say a hundred that I subjectively decided are
important, and compare them, but some are very similar and some
very different in the two books," he explained. "So, how can I
decide? I need a second method to compare some features
representing one whole book to those of the other whole book."
 |
|
Text comparison of English books
with the FFP method yields a relationship tree that
groups similar books together, by genre, period or
author. (Sung-Hou Kim lab/UC
Berkeley) |
A different vocabulary
Text comparison of English books with the FFP method yields a
relationship tree that groups similar books together, by genre,
period or author. (Sung-Hou Kim lab/UC Berkeley)Teaming up with
biophysicist Gregory E. Sims, statistical mathematician Se-Ran
Jun and theoretical physicist Guohong A. Wu, Kim decided to try
a simple variant of the word frequency technique. They
eliminated all punctuation and spaces from a text, created a
dictionary of all the two-letter, three-letter, and other word
combinations in the books, and counted the variety of each
fixed-length "word" or feature. The features were not
consecutive letter combinations, but overlapping sequences
obtained by sliding a two-, three- or more-letter window along
the text, advancing one letter at a time.
In a test of free online books obtained through Project
Gutenberg, they found that this method, which they called the
feature frequency profile (FFP) method, was more successful at
identifying related books - books by the same author, books of
the same genre, books from the same historical era - than word
frequency profile analysis. In fact, a good tree can be
constructed by looking at a single "optimal" feature length,
such as nine letters, where the "vocabulary" is very large,
instead of looking at all possible lengths.
"I was just stunned when I saw this," Kim said. One of the
reasons this method works better, he said, may be that, while
word frequency analysis treats each word independently, feature
frequency analysis picks up syntax.
"Here, if I take a 9-letter window and slide it along the text,"
he said, "I am actually picking up the relationship between the
first and second words - the local syntax - which was impossible
to pick up from the word frequency method. Apparently, that is
very important."
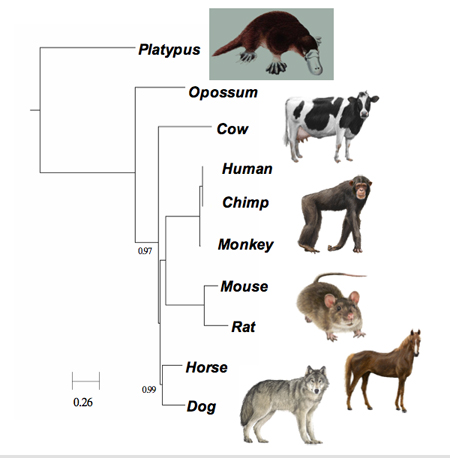 |
|
FFP comparison of
mammalian genomes produces the same phylogenetic tree
whether using whole genomes or just introns, which
supposedly carry no genetic information. (Sung-Hou Kim
lab/UC Berkeley) |
Mammalian and bacterial genomes
Buoyed by this success, the researchers applied the technique to
whole genomes of mammals, where there is the least controversy
in evolutionary relationship. "We treat the genome like a book
without spaces," Kim said.
FFP comparison of mammalian genomes produces the same
phylogenetic tree whether using whole genomes or just introns,
which supposedly carry no genetic information. (Sung-Hou Kim
lab/UC Berkeley)Since these genomes are very large, the
researchers translated the genome sequences using a reduced,
two-letter alphabet - R for the purine nucleic acids, adenine
and guanine, and Y for the pyrimidine nucleic acids, thymine and
cytosine - to reduce the complexity of calculation. Using an
optimal feature length of 18 base pairs, this test created a
family tree identical to the phylogenetic trees constructed by
scientists using genetic, morphological, anatomical, fossil and
behavioral information. This was surprising, especially since
the overwhelming majority of the mammalian genomes do not code
for genes, Kim said.
Next, they tried the FFP method on 518 genomes, the bulk of them
bacteria and Archaea, but also six eukaryotes of varying
complexity and two random sequences. The eurkaryotic genomes
used were as much as 1,000 times longer than the bacterial and
Archaeal genomes. Because most of the bacterial and Archaeal
genomes code for genes, as opposed to very little of the genomes
of higher eukaryotes, the researchers used a different alphabet
and vocabulary for the FFP method: short strings of amino acids,
the building blocks of proteins, with a 20-word alphabet
representing the 20 possible amino acids.
"The question is: Can we then group all living organisms based
on the whole proteome, that is, the assembly of all proteins,
instead of using just a selection of a small set of proteins,
which is equivalent to using a small set of genes?" said Kim.
The researchers found that the FFP method clearly segregates
whole proteomes of all bacteria, archaea, eukaryotes and random
sequences into separate groups or domains. Most of the phylum
groups within each domain and class groups in each phylum also
were well segregated, with some interesting discrepancies
compared to the currently accepted groupings.
In most of the cases where the FFP grouping disagreed with an
accepted phylogenetic grouping, the problem organism had been
the subject of debate among biologists because of conflicting
conclusions from genetics, behavior and morphology, Kim said.
The new method did classify several so-far unclassified
bacteria, however.
The major differences are found not in how the organisms are
grouped, but in the relative position of these groups in the
organism trees, he said.
Viral genomes
Finally, Kim and his colleagues analyzed the genomes of several
hundred viruses, including several that could not be classified.
"Some viruses have no or few highly conserved common genes to
other viruses, thus, the gene alignment-based method cannot find
relationship among such groups, but we think we can," he said.
Because of the vast amount of whole genome sequence data, all of
Kim's analyses monopolized a computer cluster of 320 CPUs
(central processing units) for over a year.
Kim stressed the major difference between FFP and gene-centric
comparisons of genomes: FFP takes into account all or most of
the DNA or protein sequences in the genome, while gene alignment
analysis chooses a small set of genes out of large number of
genes in each organisms, and uses that to represent the
organism.
"The fallacy of the view that organisms can be represented by a
small set of their genes is really due to our prejudice that
genes are us," Kim said. "We know now, more and more, that this
is oversimplification.
"It is likely that some of the observations we come up with will
turn out to be wrong, but the method will evolve and get better
and better as experts come in and tell us where we have gone
wrong. The math is there, now we have to remove the human bias
as much as possible."
In addition to applying the method to comparative genomics, Kim
expects it will help in grouping and finding relationships among
sets of other information, such as electronic information
encoding text, sounds and images. It may also help in tracing
human ancestry and disease demography using whole genome
sequences, and in grouping of metagenomic data - the sequences
of genome fragments from many organisms, most of which are
unknown species, found in a given environmental niche or body
organ.
Kim hopes someday to return to Shakespearean texts and sort out
their provenance as well.
The work was funded by the National Institutes of Health and by
a grant from the Korean Ministry of Education, Science and
Technology.
By Robert Sanders, Media Relations |
|