|
June 2003
from:
Keygene Genetics
Newsletter VI
Linkage
Disequilibrium Mapping (LD Mapping), allows for testing
associations between phenotype and DNA markers directly on
germplasm lines (instead of segregating populations).
A
prerequisite to execute this approach is the availability of a
database with many thousands of genetic markers for which the
precise map position is known. To date, AFLP is the only
technology which can generate these numbers of marker within a
reasonable amount of time and effort. Efficient mapping of the
thousands of markers can be achieved using a BAC pooling
strategy.

Ultra
High Density Mapping using BAC Pools of Pristionchus pacificus,
a free living nematode. The vast majority of bands from the
parent source segregate in the BAC pools, and this segregation
pattern is used to physically map the marker bands. S: size
standard; P: parental source from which the BAC library is
constructed.
As time
goes by, the number of agronomically important species for which
the complete genome is sequenced will increase. By analyzing the
genome sequence, the length of AFLP fragments for each AFLP
primer combination can be predicted. Vice-versa, for each band
in an AFLP fingerprint of the sequenced sample, the genome
position can be traced back through the genome sequence.
Keygene has
built a software package called REcomb which can map AFLP bands
in silico
using the
complete genome sequence. Together with the AFLP technology,
this tool allows us to efficiently generate a germplasm dataset
containing many thousands of mapped markers, the ideal start for
successful LD mapping work.
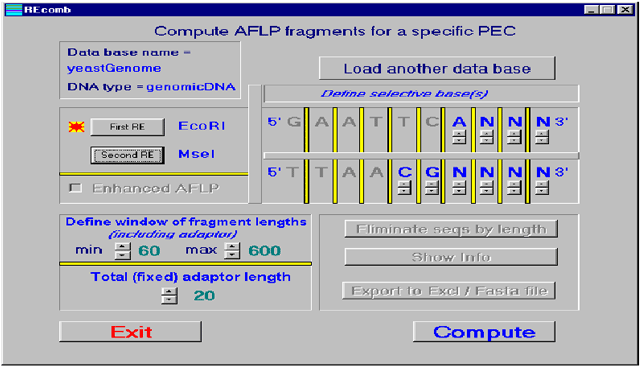
Example
of how our REcomb software enables the calculation of all
expected AFLP fragments which will be derived from a total
genome for specific Restriction Enzyme combinations (and
possibly also primer extensions). |