Washington, DC
February 28, 2008
A consortium of researchers led by
the Genome Sequencing Center (GSC) at Washington University in
St. Louis, Missouri, announced today the completion of a draft
sequence of the corn genome.
In the fall of 2005 the National
Science Foundation (NSF), in partnership with the U.S.
Department of Agriculture (USDA) and the Department of Energy
(DOE), awarded $32 million to two projects to sequence the corn
genome. The goal of the project led by the Washington University
GSC is to develop a map-based genome sequence for the B73 inbred
line of corn.
This groundbreaking sequencing project was funded by the NSF
under the auspices of the National Plant Genome Initiative
(NPGI). The initiative, which began in 1998, is an ongoing
effort to understand the structure and function of all plant
genes at levels from the molecular and organismal, to
interactions within ecosystems. NPGI's focus is on plants of
economic importance and plant processes of potential economic
value. Sequencing the corn genome is one of the major goals of
the current initiative.
"Corn is one of the most economically important crops for our
nation," said NSF Director, Arden L. Bement, Jr. "Completing
this draft sequence of the corn genome constitutes a significant
scientific advance and will foster growth for the agricultural
community and the economy as a whole."
According to the USDA, more than 80 million acres of land in the
United States is devoted to growing corn, accounting for more
than 90 percent of the total value of feed grain.
"Corn is a vitally important crop," said Rick Wilson, lead
investigator and director of the GSC. "Scientists will now be
able to accurately and efficiently probe the genome to develop
new varieties of corn that increase crop yields and resist
drought and disease. The information we glean from the corn
genome is also likely to be applicable to other grains, such as
rice, wheat and barley."
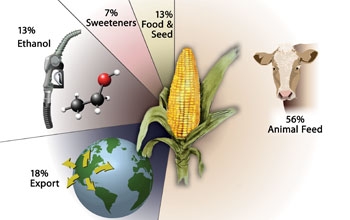 |
|
Corn, also
known as maize, is used to produce a myriad
of products, from breakfast cereal, meat and
milk to toothpaste, shoe polish and ethanol.
Credit: Nicolle Rager Fuller, National
Science Foundation |
|
Sequencing the corn genome has been an immense and daunting
task. At 2.5 billion base pairs covering 10 chromosomes, this
genome's size is comparable to that of the human genome. Corn
also has one of the most complex genomes of any known organism
and is one of the most challenging genomes sequenced to date.
The draft sequence will allow researchers to begin to uncover
the functional components of individual genes as well as develop
an overall picture of the genome organization. Completing the
draft sequence, which covers about 95 percent of the genome, is
an important milestone on the way to refining the complete
genome sequence.
"Creating a completed draft of the corn genome brings us one
step closer to our goal of understanding the functional genetic
components that influence hybrid vigor, drought and pest
resistance, and asexual plant reproduction or apomixis - all
special traits that make corn valuable," said James Collins,
head of the Biological Sciences Directorate at the NSF.
The National Corn Growers Association, a strong supporter of the
sequencing project and an advocate of the NPGI, notes that
elucidating the complete sequence and structure of all corn
genes, associated functional sequences and their locations on
corn's genetic and physical map, has many potential benefits.
These include: creating a model for other major genome
sequencing projects, enhancing the efficiency of modern corn
breeding programs, increasing understanding of corn's important
agronomic traits, and strengthening the physical and
intellectual scientific processes of the genetic research
community.
Pam Johnson, chairman of the Research and Business Development
Action Team for the National Corn Growers Association, adds,
"This effort is especially critical at this time in history,
when the growing global population looks to corn and other
plants to supply food, feed, bioenergy and biobased materials.
It is time to learn the language of corn as a model that has
great potential and economic significance."
Collaborators contributing to the GSC corn genome research
include: Rod Wing from the University of Arizona; W. Richard
McCombie, Robert Martienssen, Doreen Ware, and Lincoln Stein
from Cold Springs Harbor Laboratory; Patrick Schnable and
Srinivas Aluru from Iowa State University; and Richard Wilson
and Sandy Clifton from Washington University.
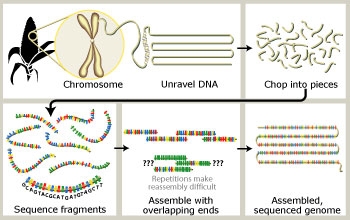 |
|
To sequence
the maize genome, scientists first collect
and purify DNA from maize plants in the
laboratory. Purified DNA is "chopped up" to
produce DNA small enough to analyze. A
sequencing machine determines the actual
order of about 1,000 DNA bases (abbreviated
G, A, T, or C) at a time. By analyzing the
sequence data with sophisticated computer
programs, the fragments can be aligned by
overlapping their ends. Repeated sequences
throughout the genome make it difficult to
match up the correct pieces. When the
project is completed, researchers will know
the sequence of all 2.5 billion DNA bases in
the maize genome.
Credit: Nicolle Rager Fuller, National
Science Foundation |
|
The National Science Foundation (NSF) is an independent federal
agency that supports fundamental research and education across
all fields of science and engineering, with an annual budget of
$5.92 billion. NSF funds reach all 50 states through grants to
over 1,700 universities and institutions. Each year, NSF
receives about 42,000 competitive requests for funding, and
makes over 10,000 new funding awards. The NSF also awards over
$400 million in professional and service contracts yearly. |
|
|
|
 |
|
In
2005, NSF, DOE, and USDA funded
the sequencing of the corn
genome. Researchers for the
sequencing project are from
Washington University, Cold
Spring Harbor Laboratory, Iowa
State University, and the
University of Arizona.
Credit: © 2008 Jupiter Images
Corp. |
|
|
|