|
Ithaca, New York
October 24, 2002
 |
|
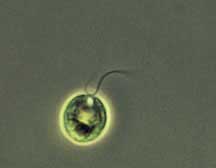
Using
a phase-contrast microscope, the plant Chlamydomonas
is magnified about 1000 times.
Credit: Yoshiki Nishimura/Boyce Thompson Institute -
Copyright © Cornell University |
With the genomes of humans and
several insects, animals and crop plants mapped or sequenced,
biologists are turning their attention to single-celled algae no
thicker than a human hair. Among the possible payoffs: crops
requiring less fertilizer, a source of renewable energy and a
new source for novel proteins.
The algae, Chlamydomonas
reinhardtii , already are an important biological model
for genetics research. Now, the complete genome of the plant's
chloroplast has been sequenced by scientists at the
Boyce Thompson Institute
(BTI) for Plant Research located on the campus of Cornell
University. The chloroplast is the area of the plant that
harvests light energy. Details of the sequencing (that is,
determining the base sequence of each of the ordered DNA
fragments) appear in the latest issue of the journal The
Plant Cell (November 2002).
The complete chloroplast genome
sequence, says David Stern, a biologist and vice president for
research at BTI, a not-for-profit research organization, has
made it possible to test the response of Chlamydomonas
(pronounced CLAMMY-doe-moan-us) to various environmental
stresses, work that is reported in an accompanying article in
The Plant Cell . In addition, the organism's nuclear genome
is being sequenced by the Joint Genome Institute, a unit of the
Department of Energy.
One type of environmental stress
being explored, says Stern (who also is an adjunct professor of
plant biology at Cornell) is that of response to phosphates. The
developed world, he says, puts too much phosphorous fertilizer
on plants and crops. "It turns out that Chlamydomonas shares
many of the responses to phosphate stress with crop plants.
Working with Chlamydomonas, we can quickly test ways to improve
tolerance or adaptation, perhaps leading to ways of engineering
crop plants for the same purpose," he says.
If fertilizer use were decreased,
phosphorous runoff into creeks, streams and lakes might be
diminished. Phosphate leaching is a prime cause of algae blooms
in lakes and ponds around agricultural areas.
The algae might also one day be a
source of hydrogen, a clean-burning fuel. At present, hydrogen
used in a type of battery called a fuel cell (still in its
infancy for powering cars and boats) is extracted from natural
gas -- a nonrenewable resource. A group led by Anastasios Melis,
a professor at the University of California-Berkeley, is
exploring the use of Chlamydomonas as a renewable hydrogen
source.
Additional applications include
using the Chlamydomonas chloroplast as a "bioreactor" to create,
or "over-express," a variety of novel proteins for agricultural,
industrial and biomedical purposes, says Jason Lilly, a BTI
postdoctoral researcher.
Chlamydomonas plants have been
useful to science for a century in both agriculture and energy
research. In nature, the organisms are widely present in fresh
and brackish water, all kinds of soils, underwater thermal vents
and even under the Antarctic ice shelf. One species of
Chlamydomonas sports a red pigment -- as protection from solar
damage -- and is found in alpine or arctic regions. These red
algae create a phenomenon referred to as "red snow." The
organism's global dispersion demonstrates the algae's adaptive
nature, says Stern.
"Chlamydomonas is a relatively
simple organism and easy to work with," says Stern. "One
drawback, however, is that despite its long history as a
laboratory organism, the scientific community has lacked
so-called genomics resources. This long-awaited part of the
genetic toolbox promises to be a boon for scientists."
Completing the Chlamydomonas
chloroplast genome sequence is part of a larger Chlamydomonas
Genomic Initiative, spearheaded by Arthur Grossman of Stanford
University, working at the Carnegie Institution of Washington.
Stern's chloroplast genome studies, a project that began three
years ago, have been supported by grants from the National
Science Foundation and the National Institutes of Health.
In addition to Stern and Lilly,
the other authors and researchers are: Jude E. Maul, BTI
laboratory researcher; Liying Cui, Claude W. dePamphilis and
Webb Miller of Pennsylvania State University; and Elizabeth
Harris of Duke University. The articles, "Chlamydomonas reinhardtii
plastid chromosome: Islands of genes in a sea of repeats"
and "The Chlamydomonas reinhardtii organellar
genomes respond transcriptionally and post-transcriptionally to
abiotic stimuli," are scheduled to appear in the November
printed issue and the online edition of The Plant Cell .
Related World Wide Web sites
The following sites provide additional information on this news
release:
- Chloroplast Genome web site at
the Boyce Thompson Institute:
bti.cornell.edu/bti2/chlamyweb/
- The Chlamydomonas Genomic
Project:
http://www.biology.duke.edu/chlamy_genome/index.html
- Joint Genome Institute:
http://bahama.jgi-psf.org/prod/bin/chlamy/home.chlamy.cgi
|